kinGEMs: Enzyme-Constrained Genome-Scale Models Pipeline
A pipeline for constructing enzyme-constrained genome-scale models with deep learning predicted kinetic parameters.
kinGEMs: Enzyme-Constrained Genome-Scale Models Pipeline
Overview
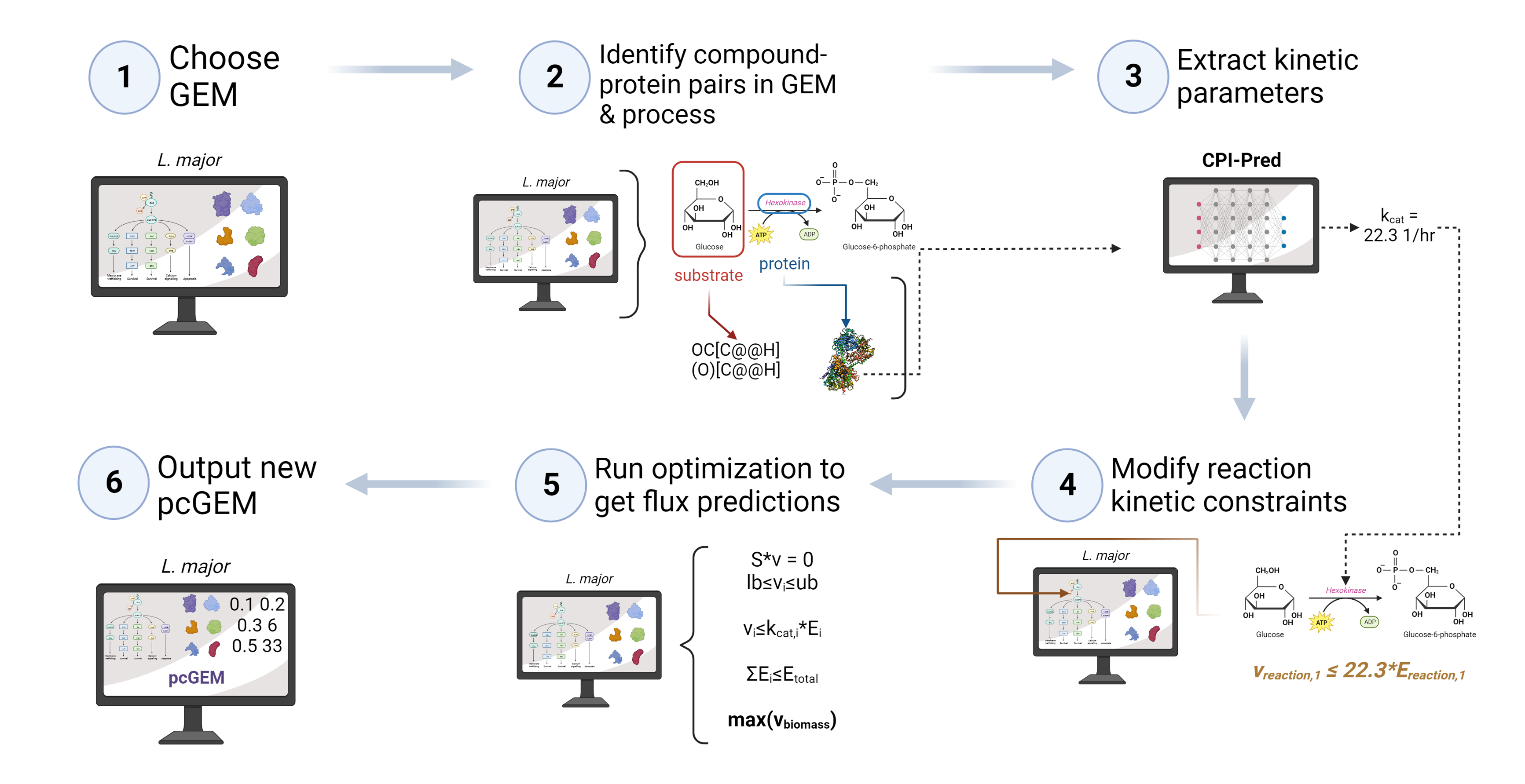
The kinGEMs pipeline represents a breakthrough in genome-scale modeling by integrating enzyme constraints with deep learning-predicted kinetic parameters, enhancing the biological realism of GEMs. KinGEMs constructs enzyme-constrained genome-scale models (ecGEMs) using CPI-Pred, a custom deep learning model trained to predict kcat, KI, and KM values for enzyme-substrate pairs.
Key Features
- Advanced Constraint Optimization: Integrates enzyme turnover constraints directly into reaction fluxes with a flexible linear optimization framework. This setup allows for potential future incorporation of non-linear constraints, such as substrate saturation and allosteric regulation.
- Deep Learning-Driven Kinetic Predictions: CPI-Pred predicts kinetic parameters for enzyme-substrate pairs using protein sequence and substrate data extracted from the input GEM, creating organism-specific ecGEMs. This includes predictions for kcat, KI, and KM values, trained on one of the largest kinetic parameter datasets (~45k kcat, ~85k KM, and ~77k KI datapoints).
- Application Across Diverse Organisms: kinGEMs has been validated with model organisms like E. coli and S. cerevisiae and applied to non-conventional microbes, including K. lactis.
Significance
This project addresses a major bottleneck in GEM construction by providing an automated, scalable approach for incorporating enzyme constraints and kinetic parameters. kinGEMs bridges the gap between computational predictions and biological accuracy, laying the groundwork for more complex regulatory mechanisms in GEMs.
Project Components
- CPI-Pred Model: Trained on extensive kinetic data, it offers comprehensive kinetic predictions, enabling researchers to build accurate ecGEMs for various organisms. Read more about this in my other projects!
- Optimization Framework: kinGEMs uses an advanced framework that allows linear and non-linear constraint optimization, improving model flexibility and precision.
- Organism-Specific kcat Tuning: A unique feature that tunes kcat values to fit organism-specific constraints and experimental conditions, enhancing the robustness of the ecGEMs.
Further Applications
kinGEMs is versatile and adaptable, supporting advancements in metabolic engineering and synthetic biology. By generating ecGEMs with organism-specific kinetic parameters, kinGEMs enhances predictive accuracy and aids in modeling complex cellular behaviors.

GitHub Repository
Will be posted once the paper is published!