Compound-Protein Interaction Prediction
A deep learning model to predict compound-protein interactions with improved embedding techniques and transformer-inspired methods.
Compound-Protein Interaction Prediction
Overview
This project focuses on predicting interactions between compounds and proteins using advanced machine learning models. By leveraging protein language models and custom embedding pooling techniques, we enhance accuracy in function prediction tasks over traditional methods.
Methodology
- Protein Encoding: Various protein language models with advanced embedding pooling methods were employed to capture function-specific information, improving prediction performance.
- Compound Encoding: My colleague Zhiqing Xu developed a novel directed message-passing neural network (d-MPNN) using Extended Connectivity Fingerprints (ECFP) to represent compounds.
- Interaction Simulation: Transformer-inspired architectures model the interaction between proteins and compounds, enabling accurate prediction across multiple kinetic parameters.
Key Contributions
- Demonstrated improved function prediction (especially in kinetic parameter prediction) with advanced pooling methods.
- Introduced transformer-inspired architectures for better compound-protein interaction simulation.
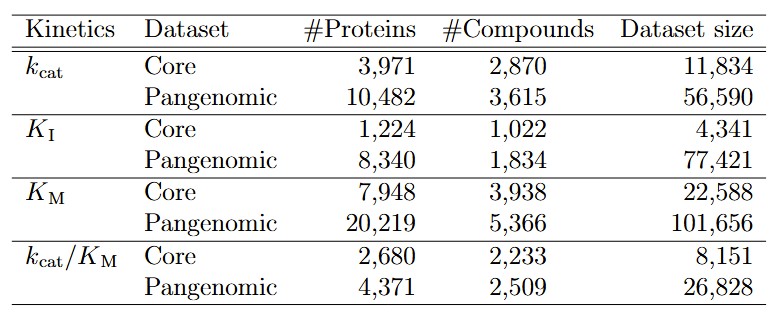
- Developed machine-learning-ready kinetic datasets for benchmarking sequence-to-function models.
Further Information
This project provides a robust framework for compound-protein interaction prediction, aiding research in drug discovery, enzyme engineering, and other biotechnology applications. The datasets created are accessible for benchmarking in related research.


GitHub Repository
Will be posted once the publication is out! Please stay tuned.