KinMod: Metabolism Regulation Database
KinMod is a tool that can be used for investigating large-scale metabolism and the regulatory mechanisms involved.
KinMod Database
Overview
KinMod is an open-access database created to catalog and analyze regulatory interactions in metabolic networks. This tool provides a curated dataset on enzyme modifications and their effects on metabolic pathways, supporting metabolic engineering and synthetic biology research.
Objectives
- Aggregate enzyme regulation data across various organisms.
- Offer insights into enzyme kinetics and regulatory interactions to aid pathway design.
- Provide customizable search options for researchers and metabolic engineers.
Key Features
- User-Friendly Interface: Allows intuitive exploration of enzyme regulatory data.
- Data Integration: Cross-referenced with other biological databases for comprehensive information.
- Customizable Searches: Users can search by enzyme, organism, or regulatory modification type.
Further Reading
For a more detailed description of KinMod, refer to our publication:
“KinMod: Metabolism Regulation Database”, Database, Oxford University Press and GitHub repo: LMSE/KinMod.
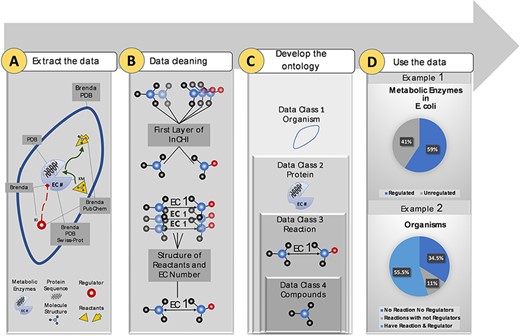
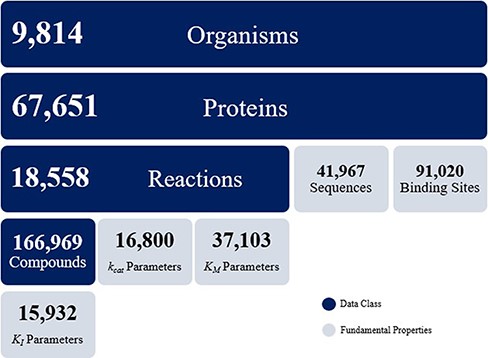

Above: KinMod's data integration, data summary, and search functionalities supporting metabolic research.